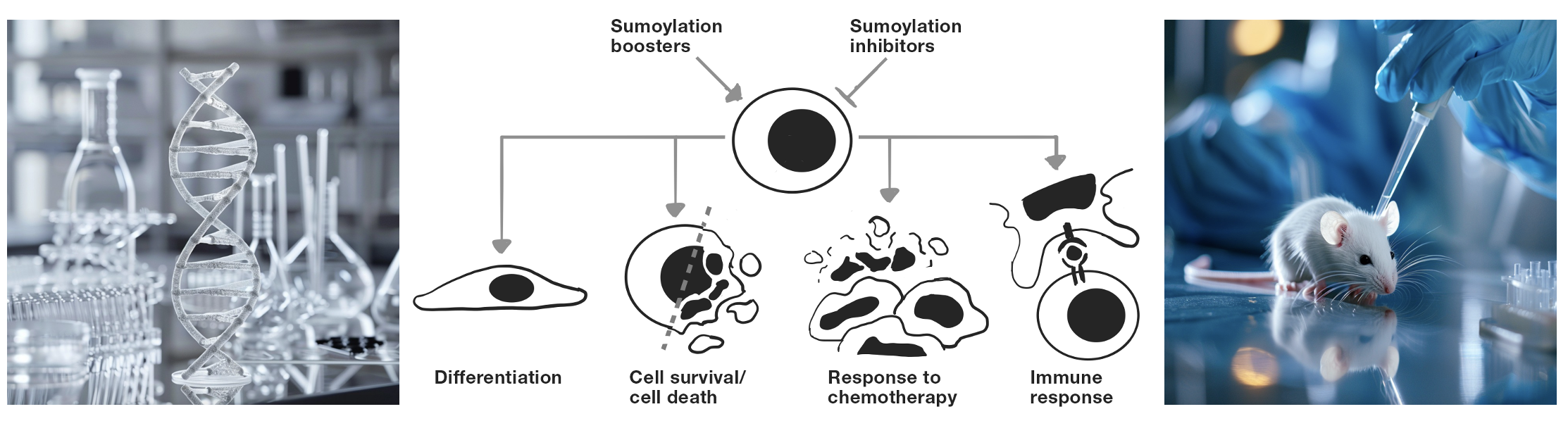
Our lab is interested in studying the biology of SUMO (Small ubiqutin-like modifier) proteins, their role in pathogenesis, as well as their potential for the development of targeted therapies. SUMO is a small, ubiquitin-like protein that is highly conserved and can be attached to various protein substrates, expanding the functional diversity of the eukaryotic proteome. This process, known as SUMOylation, is a crucial post-translational modification, evolved to accomodate some of the complexities of eukaryotic cells and to regulate the intricate networks within these systems.

Thousands of eukaryotic proteins are subject to and modified by SUMOylation, leading to changes in their structure, function, localization, stability or interactor profile. At the cellular level, SUMOylation is a key regulator of nuclear and genomic integrity, transcription, cell proliferation, stemness, senescence and infection, as well as of epigenetic and metabolic processes. At the physiological level, it acts as a crucial regulator of development. The involvement of SUMOs in a wide variety of cellular mechanisms has shaped our understanding of many pathological conditions, including cancer, neurodegeneration, and infectious diseases.

We conduct curiosity-driven fundamental research to dissect the functions of SUMOylation in key cellular processes and pathogenesis, with a major interest in disease biology and drug discovery. To achieve this, we employ multidisciplinary approaches, including cell biology, biochemistry, high-resolution imaging, proteomics, and transgenic animal models of disease.

For additional details, please check out our recent reviews:
- Sumoylation on its 25th anniversary: mechanisms, pathology, and emerging concepts – Celen and Sahin, FEBS Journal (2020) – PubMed
- Sumoylation in Physiology, Pathology and Therapy – Sahin, de Thé, Lallemand-Breitenbach, Cells (2022) – PubMed
In collaboration with IPSEN Pharma, we recently reported a novel molecule, IRC117539, that enhances the degradation of the androgen receptor (AR), a key oncoprotein in prostate cancer, by promoting its SUMOylation and ubiquitylation. This mechanism reduces prostate cancer cell viability by inhibiting AR signaling, including in androgen-insensitive cells, a critical issue in clinical cases of castration-resistant prostate cancer. In xenograft mouse models, IRC117539 showed tumor suppression efficiency comparable to enzalutamide. Our study underscores the potential of pharmacological strategies to target oncoproteins for degradation by inducing their SUMOylation.

A growing number of key proteins are emerging as prime sumoylation targets, and we aim to define how sumoylation regulates their function. In that regard, we are very interested in the biology of the CRISPR/Cas9 system, a popular genome editing platform.
We recently reported our discovery of Cas9 sumoylation and ubiquitylation, marking the first post-translational modifications identified on this crucial enzyme. Our findings reveal that ubiquitylation leads to the proteasomal degradation of Cas9, while SUMO modification influences both the enzyme’s stability and its sgRNA-directed DNA-binding efficiency. We are currently investigating the implications of these findings for genome editing technologies. We are also interested in understanding whether Cas9 SUMOylation is an evolutionarily conserved mechanism that is consequential for host-pathogen interactions.

Indeed, SUMOylation has been shown to interact with intracellular pathogens. While SUMO can trigger the degradation of microbial proteins as a part of the innate immune response to inhibit pathogen replication during infection, some pathogens can disarm the host’s SUMOylation response or exploit it for their benefit. Our recent research demonstrated that HIV-1 disrupts cellular SUMOylation by targeting the host’s SUMO E1-activating enzyme, revealing another layer of complexity in how this virus undermines host immune responses. Upon infection, HIV-1 interferes with or takes over various cellular mechanisms and pathways, progressively harming the immune system at a physiological level. We have provided the first evidence that SUMOylation, with its crucial functions for innate immunity, is one of the cellular processes affected by HIV-1.

Our lab is particularly interested in neurodegeneration and the development of novel therapeutics for neurodegenerative diseases, specifically Amyotrophic Lateral Sclerosis (ALS). ALS is a devastating neurodegenerative disease characterized by the degeneration of motor neurons in the brainstem and spinal cord, typically leading to death within 3 to 4 years of symptom onset. It is the most common motor neuron disease in humans and the third most prevalent neurodegenerative disorder after Alzheimer’s and Parkinson’s diseases, with very limited treatment options available.
At the basic science level, we investigate the role of sumoylation in ALS pathogenesis to gain insights into the mechanisms of neurodegeneration in ALS patients. We are particularly focused on how proteins undergo liquid-liquid phase separation or evade quality control, as well as how sumoylation interacts with these processes to either induce or prevent cytotoxicity.
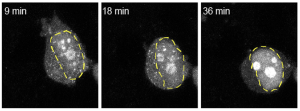
At the translational level, our long-term goal is to enhance the clearance of toxic proteins from the motor neurons of ALS patients. To this end, we are currently exploring the potential of targeting sumoylation as a therapeutic strategy.
To pursue these goals, we have recently developed a novel mouse model of ALS driven by mutant NEK1 expression and identified a crucial genetic interactor that regulates disease onset. We are characterizing this new animal model in collaboration with Hugues de Thé (Collège de France), Alain Prochiantz (Collège de France and BrainEver), and Elif Nur Firat-Karalar (Koç University). Our observations demonstrate the feasibility of SUMO-targeted protein degradation in ALS treatment and could have direct consequence in patient care (Georgiadou et al, bioRxiv, 2024).

– This website is not optimized for mobile viewing and is best experienced on desktops, laptops, or tablets –