PROJ 102 Fall 2014
 Description: The students involved in this project will examine and manipulate the 3D molecular structures of two drugs: pentamidine and dyclonine. Model construction and visualization will be performed with the software package VMD (http://www.ks.uiuc.edu/Research/vmd/).
Description: The students involved in this project will examine and manipulate the 3D molecular structures of two drugs: pentamidine and dyclonine. Model construction and visualization will be performed with the software package VMD (http://www.ks.uiuc.edu/Research/vmd/).
Then, students will examine how these drugs insert into the cell membrane. To this end, models of a fragment of the membrane and the drug molecules will be simulated in atomic detail using the molecular dynamics package NAMD (http://www.ks.uiuc.edu/Research/namd/).
Finally, students will try to connect their findings from the simulations with the experimental dose-response curves of the drugs (which are already available).
Curiosity about biological molecules and familiarity with simple programming are highly desirable for successful performance in this project
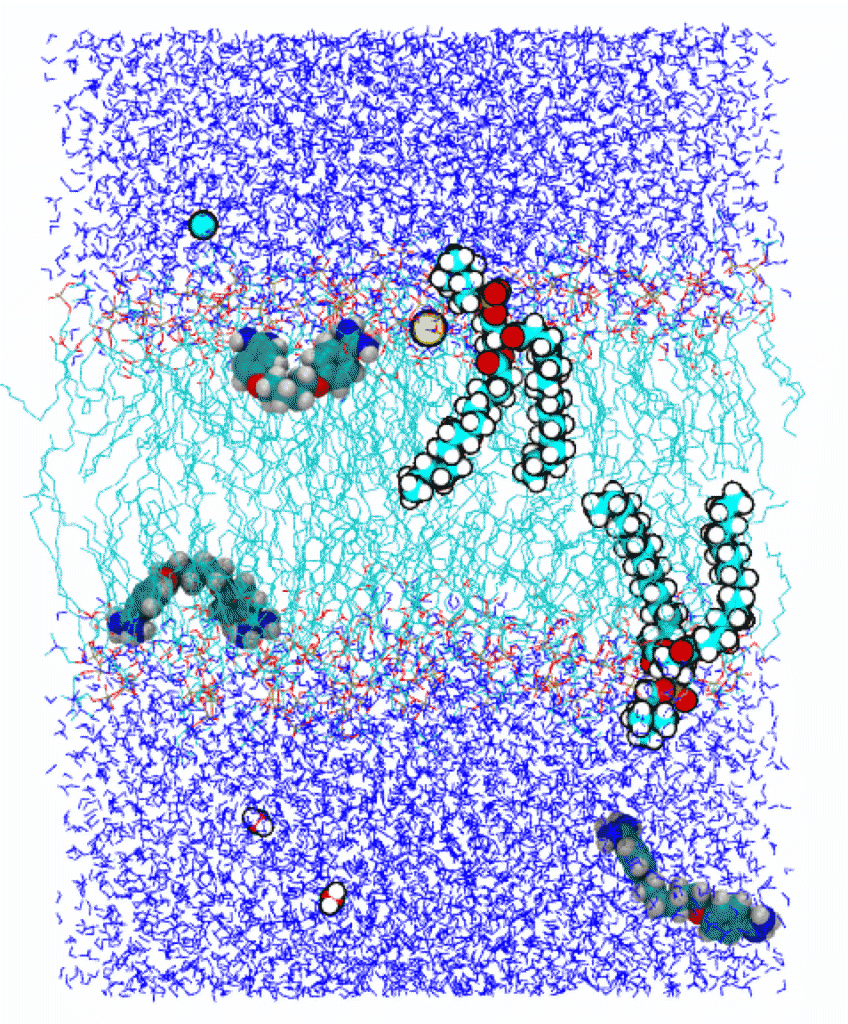
Figure: A snapshot from a molecular dynamics simulation. The lipid bilayer, in the middle, is immersed in water from both sides. (Water molecules are shown as thin, dark blue lines.) Two lipid molecules in the bilayer are shown with balls, which correspond to atoms. Oxygen atoms are colored red, nitorgen atoms are dark blue, carbon atoms are cyan, and hydrogens are white. Similarly, two water molecules, two ions, and three drug (pentamidine) molecules are represented with balls. Two pentamidines are seen to be in the bilayer while one is in water.